Note
Go to the end to download the full example code.
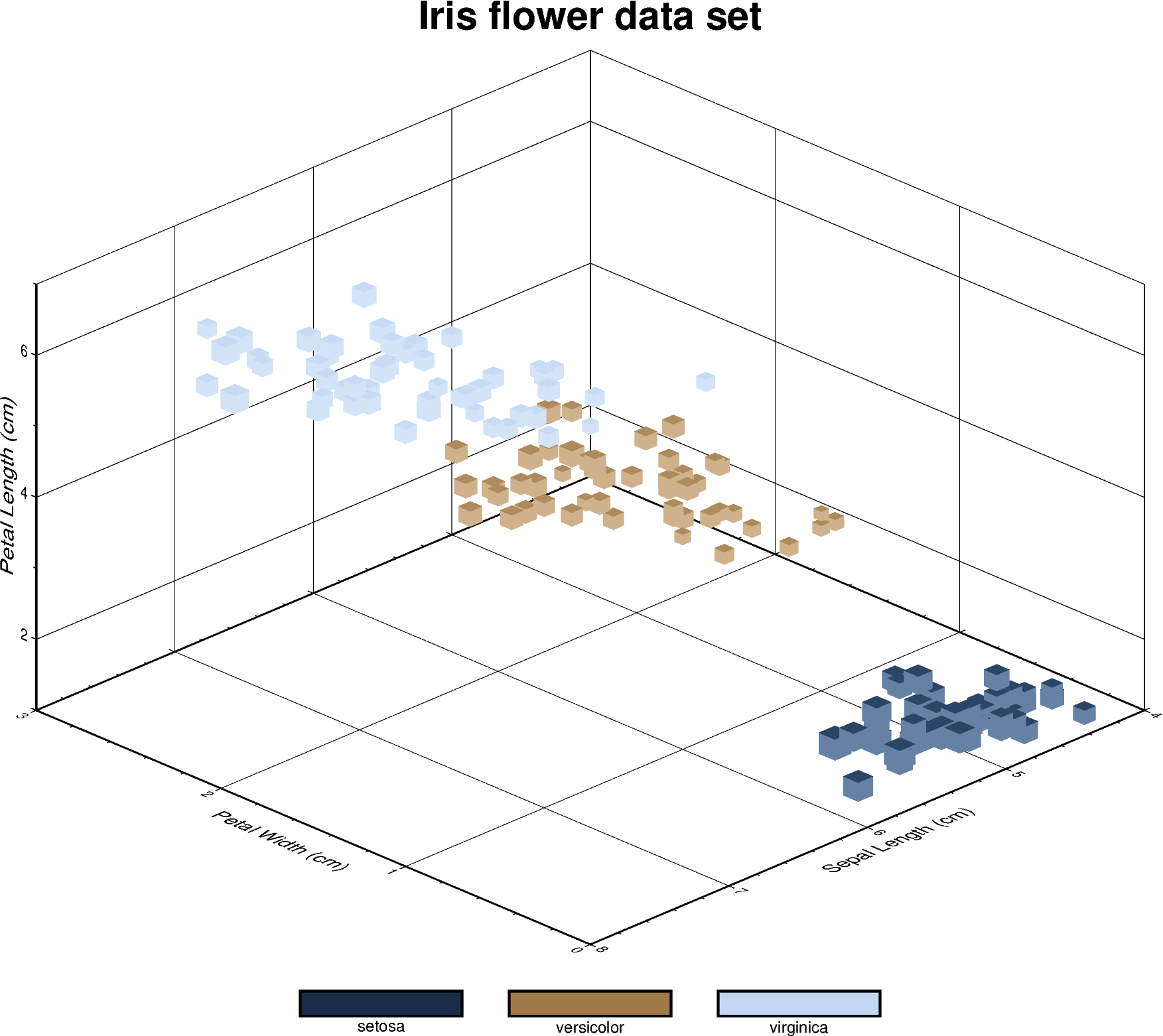
3-D scatter plots
The pygmt.Figure.plot3d method can be used to plot symbols in 3-D.
In the example below, we show how the
Iris flower dataset
can be visualized using a perspective 3-D plot. The region
parameter has to include the \(x\), \(y\), \(z\) axis limits in the
form of (xmin, xmax, ymin, ymax, zmin, zmax), which can be done automatically
using pygmt.info. To plot the z-axis frame, set frame as a
minimum to something like frame=["WsNeZ", "zaf"]. Use perspective to
control the azimuth and elevation angle of the view, and zscale to adjust
the vertical exaggeration factor.
import pandas as pd
import pygmt
# Load sample iris data
df = pd.read_csv("https://github.com/mwaskom/seaborn-data/raw/master/iris.csv")
# Convert 'species' column to categorical dtype
# By default, pandas sorts the individual categories in an alphabetical order.
# For a non-alphabetical order, you have to manually adjust the list of
# categories. For handling and manipulating categorical data in pandas,
# have a look at:
# https://pandas.pydata.org/docs/user_guide/categorical.html
df.species = df.species.astype(dtype="category")
# Make a list of the individual categories of the 'species' column
# ['setosa', 'versicolor', 'virginica']
# They are (corresponding to the categorical number code) by default in
# alphabetical order and later used for the colorbar labels
labels = list(df.species.cat.categories)
# Use pygmt.info to get region bounds (xmin, xmax, ymin, ymax, zmin, zmax)
# The below example will return a numpy array [0.0, 3.0, 4.0, 8.0, 1.0, 7.0]
region = pygmt.info(
data=df[["petal_width", "sepal_length", "petal_length"]], # x, y, z columns
per_column=True, # Report the min/max values per column as a numpy array
# Round the min/max values of the first three columns to the nearest
# multiple of 1, 2 and 0.5, respectively
spacing=(1, 2, 0.5),
)
# Make a 3-D scatter plot, coloring each of the 3 species differently
fig = pygmt.Figure()
# Define a colormap for three categories, define the range of the
# new discrete CPT using series=(lowest_value, highest_value, interval),
# use color_model="+csetosa,versicolor,virginica" to write the discrete color
# palette "cubhelix" in categorical format and add the species names as
# annotations for the colorbar
pygmt.makecpt(
cmap="cubhelix",
# Use the minimum and maximum of the categorical number code
# to set the lowest_value and the highest_value of the CPT
series=(df.species.cat.codes.min(), df.species.cat.codes.max(), 1),
# Convert ['setosa', 'versicolor', 'virginica'] to
# 'setosa,versicolor,virginica'
color_model="+c" + ",".join(labels),
)
fig.plot3d(
# Use petal width, sepal length, and petal length as x, y, and z
# data input, respectively
x=df.petal_width,
y=df.sepal_length,
z=df.petal_length,
# Vary each symbol size according to the sepal width, scaled by 0.1
size=0.1 * df.sepal_width,
# Use 3-D cubes ("u") as symbols with size in centimeters ("c")
style="uc",
# Points colored by categorical number code (refers to the species)
fill=df.species.cat.codes.astype(int),
# Use colormap created by makecpt
cmap=True,
# Set map dimensions (xmin, xmax, ymin, ymax, zmin, zmax)
region=region,
# Set frame parameters
frame=[
"WsNeZ3+tIris flower data set", # z axis label positioned on 3rd corner, add title
"xafg+lPetal Width (cm)",
"yafg+lSepal Length (cm)",
"zafg+lPetal Length (cm)",
],
# Set perspective to azimuth NorthWest (315°), at elevation 25°
perspective=[315, 25],
# Vertical exaggeration factor
zscale=1.5,
)
# Shift plot origin in x direction
fig.shift_origin(xshift="3.1c")
# Add colorbar legend
fig.colorbar()
fig.show()
Total running time of the script: (0 minutes 0.480 seconds)